| USDA-NRI Research Program | |||
| Accomplishments for Aim 2 Identify Ralstonia solanacearum race 3 biovar 2 genes involved in cold adaptation and growth in plant hosts, using a microarray-based post-genomic approach Click here to see accomplishments for Aim1 |
|
| This page contains files in PDF format. To read these files you must have Adobe Reader installed on your computer (free). | |
|
- Why is Ralstonia solanacearum Race 3 cold tolerant? Using post-genomic analysis to explore strain-specific traits (Meeting oral communication 2009). Most strains of the bacterial wilt pathogen Ralstonia solanacearum are tropical, but one group, Race 3 biovar 2 (R3b2), is adapted to cooler environments and causes disease in temperate zones and tropical highlands. We compared the growth and virulence of R. solanacearum strains GMI1000 (tropical, biovar 3) and UW551 (R3b2, temperate) at temperate and tropical temperatures. The two strains...<Read more> - Molecular responses to quantitative bacterial wilt resistance in tomato (Meeting oral communication 2009). Host resistance is the only practical control for bacterial wilt of tomato, caused by Ralstonia solanacearum, but wilt resistance is horizontal, polygenic and its basis is not understood. We found that tomato plants infected with R. solanacearum Race 3 biovar 2 strain UW551 upregulated genes in both the ethylene and salicylic acid signal transduction pathways. However, in response to...<Read more> - Comparative gene expression profile analysis of temperate and tropical strains of Ralstonia solanacearum (Meeting oral communication 2009). Bacterial wilt is a major source of crop losses on diverse hosts, primarily in warm-temperate and tropical zones. However the Race 3 biovar 2(R3bv2) subgroup of the pathogen, Ralstonia solanacearum, attacks plants in temperate zones and the highland tropics. We found that ...<Read more> - Interactions with hosts at cool temperatures, not cold tolerance, explain the unique epidemiology of Ralstonia solanacearum race 3 biovar 2 (Journal Article 2009). Most strains of the bacterial wilt pathogen Ralstonia solanacearum are tropical, but race 3 biovar 2 (R3bv2) strains can attack plants in temperate zones and tropical highlands. The basis of this distinctive ecological trait is not understood. We compared...<Read more> - Identifying gene expression differences between race 1 and race 3 strains of Ralstonia solanacearum during bacterial wilt disease development at warm and cool temperatures (Meeting Poster 2008). We designed microarrays and developed methods for extracting quality bacterial RNA from infected tomato plants to compare gene expression analysis of race 1 (GMI1000) and race 3 (UW551) strains of R. solanacearum during infection of plants at 20°C and 29°C ... <Read more> - Interactions with hosts at cool temperature, not cold tolerance, explain the unique epidemiology of Ralstonia solanacearum race 3 biovar 2 (Meeting Poster 2008). At cool temperature (20°C), R. solanacearum race 3 biovar 2 strain (UW551) was much more virulent on tomato than tropical race 1 strain (GMI1000).These data indicate interaction with plants is required for the temperate epidemiological trait of R. solanacearum race 3 biovar 2... <Read more> - Ralstonia solanacearum race 3 biovar 2 strains are not uniquely cold tolerant in vitro (Meeting Poster 2007). Two R. solanacearum strains (AW1 and K60) native to the southeast United States were more cold tolerant than race 3 biovar 2 strains (UW551 and IPO1609) at 4°C. These results challenge the assumption that the cold tolerance of R. solanacearum race 3 strains makes them a greater threat to the U. S. potato industry than native strains....<Read more> |
||
|
- Allen C, Meng F, Jacobs JM, and Babujee L. 2009. Why is Ralstonia solanacearum Race 3 cold tolerant? Using post-genomic analysis to explore strain-specific traits. Phytopathology 99 (6): S166-S166 Suppl. Abstract. Most strains of the bacterial wilt pathogen Ralstonia solanacearum are tropical, but one group, Race 3 biovar 2 (R3b2), is adapted to cooler environments and causes disease in temperate zones and tropical highlands. We compared the growth and virulence of R. solanacearum strains GMI1000 (tropical, biovar 3) and UW551 (R3b2, temperate) at temperate and tropical temperatures. The two strains grew similarly in media at 20°C and 28°C. At 28°C, both strains wilted tomato plants rapidly in a naturalistic soil-soak virulence assay. In contrast, at 20°C UW551 was much more virulent on tomato than GMI1000, suggesting that interaction with plants is required for the temperate epidemiological trait of R3b2. To understand the mechanisms of R3bv2 cold tolerance, we studied global gene expression patterns of the two strains at 20°C and 28°C using genomic microarrays. In rich medium, the strains’ expression profiles differed significantly with respect to both strain and temperature. In the four conditions, we found differential expression of genes involved in cell wall/membrane synthesis and function, carbohydrate transport and metabolism, transcription, replication, recombination and repair, and diverse unknown functions, including some specific to R3bv2. A hitherto cryptic quorum sensing system, SolIR, and a SolIR-dependent gene, aidA, were up-regulated in R3bv2 at 20°C. aidA, which was expressed around 12-fold higher at 20°C, is not present in the GMI1000 genome. Contact author: Caitilyn Allen <cza@plantpath.wisc> |
||
|
- Milling A. 2009. Molecular responses to quantitative bacterial wilt resistance in tomato. Phytopathology 99 (6): S169-S169 Suppl. Abstract. Host resistance is the only practical control for bacterial wilt of tomato, caused by Ralstonia solanacearum, but wilt resistance is horizontal, polygenic and its basis is not understood. We found that tomato plants infected with R. solanacearum Race 3 biovar 2 strain UW551 upregulated genes in both the ethylene and salicylic acid signal transduction pathways. However, in response to R. solanacearum infection the horizontally wilt-resistant tomato line H7996 activated expression of these defense genes faster and to a greater degree than did susceptible cultivar Bonny Best. Interestingly, results suggest different roles for the virulence factor extracellular polysaccharide (EPS) in resistant and susceptible hosts. Wild-type UW551 induced a lower defense response in susceptible tomato than did an eps mutant, supporting the idea that EPS can shield R. solanacearum from recognition. In contrast, the eps mutant induced significantly lower defense responses in resistant H7996 than the wild-type strain, weakening the “cloaking” hypothesis. The eps mutant also induced noticeably less accumulation of the defensive reactive oxygen species hydrogen peroxide in resistant tomato leaves, despite attaining similar cell densities in planta. Further, cell-free purified EPS from UW551 triggered significant defense gene expression in resistant but not in susceptible tomato plants. Collectively, these data suggest that H7996 specifically recognizes EPS from R. solanacearum. Contact author: Annet Milling <cza@plantpath.wisc> |
||
|
- Jacobs JM, Meng F, Babujee L, and Allen C. 2009. Comparative gene expression profile analysis of temperate and tropical strains of Ralstonia solanacearum. Phytopathology 99 (6): S57-S57 Suppl. Abstract. Bacterial wilt is a major source of crop losses on diverse hosts, primarily in warm-temperate and tropical zones. However the Race 3 biovar 2(R3bv2) subgroup of the pathogen, Ralstonia solanacearum, attacks plants in temperate zones and the highland tropics. We found that a phylotype II R3bv2 strain, UW551, and a phylotype I tropical strain, GMI1000, grew equally well in culture at tropical and temperate temperatures and caused comparable wilting on tomato at tropical temperatures. In contrast, at a temperate 20C, the R3bv2 strain was a much more aggressive pathogen. Although a large core of ORFs are conserved across R. solanacearum strains, about 10% of the R3bv2 ORFs are not present in the GMI1000 genome; the functions of these genes may explain the biological differences between the strains. We designed whole-genome microarray chips for UW551 and GMI1000 to test the hypothesis that the strains express different transcriptomes at different temperatures in culture and during pathogenesis. Multiple familiar factors contribute to R. solanacearum virulence, but little is known about the specific functions this pathogen needs to succeed in its understudied habitat, the plant xylem tissue. Characterization of plant-induced and low-temperature-induced genes should identify mechanisms underlying both wilt pathogenesis in general and the specific temperate ecological trait of R3bv2. Contact author: Caitilyn Allen <cza@plantpath.wisc> |
||
|
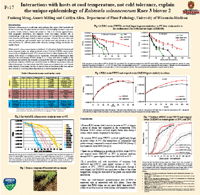
- Milling, A., Meng, F., Denny, T. P., and Allen, C. 2009. Interactions with hosts at cool temperatures, not cold tolerance, explain the unique epidemiology of Ralstonia solanacearum race 3 biovar 2. Phytopathology 99:1127-1134. Abstract. Most strains of the bacterial wilt pathogen Ralstonia solanacearum are tropical, but race 3 biovar 2 (R3bv2) strains can attack plants in temperate zones and tropical highlands. The basis of this distinctive ecological trait is not understood. We compared the survival of tropical, R3bv2, and warm-temperate North American strains of R. solanacearum under different conditions. In water at 4 degrees C, North American strains remained culturable the longest (up to 90 days), whereas tropical strains remained culturable for the shortest time (approximate to 40 days). However, live/dead staining indicated that cells of representative strains remained viable for >160 days. In contrast, inside potato tubers, R3bv2 strain UW551 survived >4 months at 4 degrees C, whereas North American strain K60 and tropical strain GMI1000 were undetectable after <70 days in tubers. GMI1000 and UW551 grew similarly in minimal medium at 20 and 28 degrees C and, although both strains wilted tomato plants rapidly at 28 degrees C, UW551 was much more virulent at 20 degrees C, killing all inoculated plants under conditions where GMI100 killed just over half. Thus, differences among the strains in the absence of a plant host were not predictive of their behavior in planta at cooler temperatures. These data indicate that interaction with plants is required for expression of the temperate epidemiological trait of R3bv2. Contact author: Caitilyn Allen <cza@plantpath.wisc> |
||
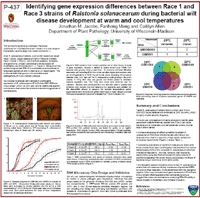
Jacobs JM, Meng F, and Allen C. 2008. Identifying gene expression differences between race 1 and race 3 strains of Ralstonia solanacearum during bacterial wilt disease development at warm and cool temperatures. Phytopathology 98 (6): S73-S73 Suppl.
Abstract: The plant pathogenic bacterium Ralstonia solanacearum elicits wilt disease on many hosts, causing significant losses for farmers worldwide. One group in this species complex, race 3 biovar 2 (R3bv2), persists and causes disease at moderately cool temperatures, in contrast to tropical R. solanacearum strains. Genome sequences of R3bv2 strain UW551 and tropical Race 1 biovar 3 strain GMI1000 suggest about 10% of UW551 ORFs are not present in GMI1000; these may explain R3bv2’s temperate epidemiology. UW551 and GMI1000 behave similarly in vitro at cold (4°C), cool (20°C) and warm (29°C) temperatures, but UW551 is nevertheless much more virulent on tomato plants at 20°C than GMI1000. This result suggests that we must study UW551 gene expression in planta to understand how it causes wilt disease at cooler temperatures. These phenotypic observations and the genomic differences between strains will frame a comparative gene expression analysis of GMI1000 and UW551 during infection of tomato plants at 20°C and 29°C. We therefore designed microarrays representing the GMI1000 and UW551 genomes and developed methods for extracting quality bacterial RNA from infected tomato plants. This powerful four-way comparison of two different strains and temperatures should suggest the molecular mechanisms that govern R3bv2 cold tolerance. Contact author: Caitilyn Allen <cza@plantpath.wisc> |
||
Meng F, Milling A, and Allen C. Interactions with hosts at cool temperature, not cold tolerance, explain
the unique epidemiology of Ralstonia solanacearum race 3 biovar 2. Phytopathology 98 (6): S104-S104 Suppl.
Abstract: Most strains of the bacterial wilt pathogen Ralstonia solanacearum are tropical, but one group, R3bv2, can attack plants in temperate zones and highland tropics. Extensive epidemiological data document the destructiveness of R3bv2 in cooler climates, but the basis of this distinctive ecological trait is not understood. We compared the survival, growth and virulence of two R. solanacearum strains, GMI1000 (race 1, tropical) and UW551 (R3bv2, temperate) at different temperatures with and without host plants. At 4°C (the temperature of commercial seed potato storage), neither GMI1000 nor UW551 survived more than 90 days in water. However, UW551 survived more than 4 months in inoculated potato tubers at 4°C, while GMI1000 survived less than 50 days in tubers. The two strains grew similarly in minimal media at 20°C and 29°C. At 29°C, both strains wilted tomato plants rapidly in a naturalistic soil-soak virulence assay. In contrast, at 20°C UW551 was much more virulent on tomato than GMI1000. Thus, there was little difference in growth and survival of tropical and R3bv2 strains at 4°C, 20°C, or 29°C in the absence of a plant host. But at cooler temperatures R3bv2 survived longer in tubers and caused disease better than a tropical strain. These data indicate interaction with plants is required for the temperate epidemiological trait of R3bv2. Contact author: Caitilyn Allen <cza@plantpath.wisc> |
||
Denny TP, Milling AS, Bhakta VG, and Allen C. 2007. Ralstonia solanacearum race 3 biovar 2 strains are not uniquely cold tolerant in vitro Phytopathology 97 (7): S28-S28 Suppl.
Abstract: The Ralstonia solanacearum race 3 biovar 2 (R3b2) subgroup, which originated in the Andes, has been widely distributed by shipment of latently infected potato tubers and can survive and cause potato brown rot in cool climates. However, R3b2 is not present in North America and APHIS listed it as a Select Agent because its supposed cold tolerance potentially threatens the potato industry. We found that R. solanacearum strains vary in their ability to survive exposure to 4°C in pure water. Contrary to expectations, strains AW1 and K60, which are native to the southeastern United States, remained culturable for more than 10 weeks and were the most cold tolerant. GMI1000 from French Guiana was among the least tolerant, whereas R3b2 strains UW551 and IPO1609 exhibited intermediate tolerance. GMI1000 survived as well as AW1 in water at 10 to 30°C. Low concentrations of various inorganic ions, buffers and organic media components reduced the survival of both GMI1000 and AW1. Survival of AW1 in water was not strongly dependent on RpoS, a stationary phase sigma factor. A viability assay revealed that a high proportion of cells had an intact plasma membrane despite being non-culturable. Although these results must still be confirmed using more natural assays, they challenge the assumption that the cold tolerance of R3b2 strains makes them a greater threat to the U. S. potato industry than native R. solanacearum strains. Contact author: Caitilyn Allen <cza@plantpath.wisc> |
||
| Click here to see project summary - project team & collaborators - project timeline. |