Ralstonia solanacearum race 3 biovar 2
(Printer friendly version - including glossary)
Figures are not included in this version - Click here to print Figures
| Author: | Patrice G. Champoiseau of University of Florida |
| Reviewers: | Caitilyn Allen of University of Wisconsin; Jeffrey B. Jones, Carrie Harmon and Timur M. Momol of University of Florida |
| Publication date: | September 12, 2008 |
| Project title: | Ralstonia solanacearum race 3 biovar 2: Detection, exclusion, and analysis of a select agent pathogen |
| Supported by: | The United States Department of Agriculture - National Research Initiative Program (2007-2010) |
Definitions of underlined words are given in the glossary at the end of this page.
Ralstonia solanacearum race 3 biovar 2 is the plant pathogen bacterium that causes brown rot (or bacterial wilt) of potato, Southern wilt of geranium, and bacterial wilt of tomato.
R. solanacearum race 3 biovar 2 occurs in highlands in the tropics and in subtropical and some warm-temperate areas throughout the world. It has also occurred in cold-temperate regions in Europe, where several outbreaks of brown rot of potato have been reported in the last 30 years. It has been reported in more than 30 countries and almost all continents.
In the United States, several introductions of R. solanacearum race 3 biovar 2 have already occurred as a result of importation of infested geranium cuttings from off-shore production sites, but the pathogen was apparently eradicated. However, because of the risk of its possible re-introduction through importation of infected plant material, and its potential to affect potato production in cold-temperate areas in the northern United States, R. solanacearum race 3 biovar 2 is considered a serious threat to the United States potato industry. It is of quarantine importance and has been listed as a Select Agent plant pathogen under the Agricultural Bioterrorism Act of 2002.
Symptoms and signs
In potato and tomato, symptoms induced by R. solanacearum race 3 biovar 2 are very similar. At the early stages of diseases, the first visible symptoms usually appear on foliage of plants. These symptoms consist of wilting of the youngest leaves at the ends of the branches during the hottest part of the day [Photo 1a. Symptom of brown rot of potato caused by R. solanacearum showing wilting of youngest leaves of plant and Photo 1b. Symptom of bacterial wilt of tomato caused by R. solanacearum showing wilting of leaves at the end of plant branch].
At this stage, only one or half a leaflet may wilt, and plants may appear to recover at night, when the temperatures are cooler. As the disease develops under favorable conditions, the entire plant may wilt quickly and desiccate, although dried leaves remain green, leading to general wilting and yellowing of foliage and eventually plant death. Another common symptom that can be associated with bacterial wilt in the field is the stunting of plants [Photo 2a. Symptom of brown rot of potato caused by R. solanacearum showing wilting and stunting of plant and Photo 2b. Symptom of bacterial wilt of tomato caused by R. solanacearum showing wilting of foliage and stunting of plant]. These symptoms may appear at any stage of plant growth.
In young stems, infected vascular bundles may become visible as long, narrow, dark brown streaks. In young, succulent plants of highly susceptible varieties, collapse of the stem may also be observed [Photo 3. Symptom of bacterial wilt of tomato caused by R. solanacearum showing collapse of young stem after artificial inoculation of the plant].
In well-established infections, cross sections of stems or stolons may reveal brown discoloration of infected tissues [Photo 4. Brown discoloration of stem tissues caused by R. solanacearum].
In potato, brown rot symptoms may be present in tubers at the later stages of disease. Cross-section of infected potato tubers may reveal a grey-brown discoloration of vascular tissues, also called a vascular ring [Photo 5. Grey-brown discoloration of vascular tissues and bacterial ooze in potato tuber infected by R. solanacearum]. As infection progresses, the discoloration may extend into the pith or cortex of the tuber. A milky-white sticky exudate (ooze), which indicates the presence of bacteria cells, might also be observed in freshly-cut sections of infected tubers [Photo 5. Grey-brown discoloration of vascular tissues and bacterial ooze in potato tuber infected by R. solanacearum].
Bacterial ooze may also be visible at the eyes or at the point where the stolon attaches to the tuber [Photo 6. Bacterial ooze exuding from eye of potato tuber infected by R. solanacearum]. These signs or symptoms may not be visible early in disease development.
In geranium, symptoms of Southern wilt usually begin with abnormal chlorosis and wilting of the lower leaves. Leaves may also show abnormal upward curling at their margins which is very characteristic of the disease [Photo 7. Initial symptoms of Southern wilt of geranium caused by R. solanacearum showing wilting and upward curling of leaves].
At this stage of disease, plants may appear to recover at night, when the temperatures are cooler. Under favorable conditions, the disease develops rapidly and wilting may spread up the plant from older leaves to newer ones. Wilted leaves often become chlorotic then brown necrotic in wedge-shaped patterns that expand towards the leaf margins. The leaf margins themselves may also become chlorotic then necrotic, and the whole plant may desiccate and die [Photo 8. Symptoms of Southern wilt of geranium caused by R. solanacearum race 3 biovar 2 showing drying and brown necrosis on leaves].
At late stages of disease, collapse of the stem may also be observed [Photo 9. Late symptoms of Southern wilt of geranium caused by R. solanacearum race 3 biovar 2 showing blackening and collapse of stem]. Stems and roots may show brown vascular discoloration, blacken and eventually become necrotic [Photo 9, Photo 10. Late symptoms of Southern wilt of geranium caused by R. solanacearum showing root blackening].
Symptom expression is favored by high temperatures (85-95°F)(29-35°C) and symptoms of the disease may progress rapidly after infection. However, under favorable conditions, symptomless plants may remain latently infected for extended periods of time. After infection the pathogen may survive in and be spread from the infected plant.
A common sign of bacterial wilt observed at the surface of freshly-cut sections from severely infected stems is a sticky, milky-white exudate, which indicates the presence of dense masses of bacterial cells in infected vascular bundles, and particularly in the xylem [Photo 11. Bacterial ooze from freshly cut section of a geranium stem infected by R. solanacearum].
Another common diagnostic sign of diseases is observed when the cut stem sections are placed in clear water as shown in Photo 12. It consists of a viscous white spontaneous slime streaming from the cut end of the stem. This streaming represents the bacterial ooze exuding from the cut ends of colonized vascular bundles [Photo 12. Bacterial streaming in clear water from stem cross-section of plant infected by R. solanacearum].
This “stem-streaming” test is easy to conduct and can be used as a valuable diagnostic tool for quick detection of bacterial wilt or brown rot in the field.
R. solanacearum race 3 biovar 2 also infects weeds and native plants, especially members of the Solanaceae. In weed hosts, wilt symptoms are rarely observed under natural conditions unless soil temperatures exceed 77ºF (25ºC) or inoculum levels are extremely high. When wilting does occur, the symptoms are as described for tomato. Solanum dulcamara (woody nightshade or bittersweet) plants growing in water may show internal discoloration of vascular tissues on the stem base without any obvious wilting. Solanum dulcamara has been described as an important weed host of R. solanacearum in England (see the causal organism section).
Causal organism
Ralstonia solanacearum (Smith 1986) Yabuuchi et al. 1996, formerly called Pseudomonas solanacearum, is a Gram-negative, rod-shaped, strictly aerobic bacterium that is 0.5-0.7 x 1.5-2.0 µm in size. It is very sensitive to desiccation and is inhibited in culture by low concentrations (2%) of sodium chloride (NaCl). For most strains, the optimal growth temperature is 82-90°F (28-32°C); however some strains that are pathogenic on potato have a lower optimal growth temperature of 80.5°F (27°C).
Liquid and solid (agar) growth media [http://plantpath.ifas.ufl.edu/rsol/Culturemedia.html] are commonly used for culture of the bacterium. On solid agar medium, individual bacterial colonies are usually visible after 36 to 48 hours of growth at 82.4°F (28°C), and two main colony types differing in morphology can be distinguished: colonies of the normal or virulent type that are white or cream-colored, irregularly-round, fluidal, and opaque; and colonies of the mutant or non-virulent type that are uniformly round, smaller, and butyrous (dry) [Photo 13. Virulent (bottom) and non-virulent (top) colonies of R. solanacearum on CPG agar growth medium].
This shift from virulent to non-virulent bacterial cells occurs during storage or under oxygen stress in liquid media. A tetrazolium chloride (TZC) medium was developed to differentiate the two colony types, on which virulent colonies appear white with pink centers and non-virulent colonies appear dark red [Photo 14. Virulent colonies of R. solanacearum on TZC agar medium]. A semi-selective medium, called modified SMSA medium, has been developed for detection of R. solanacearum in water and soil samples, and in plant extracts. On this medium, typical bacterial colonies appear fluidal, irregular in shape, and white with pink centers after 2 to 5 days incubation at 82.4ºF (28ºC) (see the detection and identification section).
For long term storage, R. solanacearum can be stored for several years at room temperature in sterilized tap, distilled or deionized water.
Many strains of R. solanacearum have been identified and characterized worldwide, revealing significant variability within the species. R. solanacearum is therefore considered a “species complex”. Based on variability in host range and in ability to utilize several carbohydrate substrates, R. solanacearum was initially subdivided into races and biovars. So far, five races and five biovars have been identified within the species [Table 1. Characteristics of races and their relationship to biovars of R. solanacearum (from Denny and Hayward, 2001; Daughtrey, 2003)], but this old classification system is unsatisfactory because it is not predictive and some groups (e.g. race 1) contain very large variation.
| Table 1. Characteristics of races and their relationship to biovars of R. solanacearum (from Denny and Hayward, 2001; Daughtrey, 2003). | |||
| Race | Primary hosts | Geographical distribution | Biovar |
| 1 2 3 4 5 |
Wide (tobacco, tomato, solanaceous and nonsolanaceous weeds, diploid bananas, groundnut, potato, pepper, eggplant, olive, ginger, strawberry, geranium, Eucalyptus, other plants…) Triploid bananas, other Musa spp. Potato and tomato Ginger Unknown Mulberry tree |
Asia, Australia, Americas Caribbean, Brazil, Philippines Worldwide except US and Canada Australia, China, Hawaii, India, Japan, Mauritius, South Asia India China |
3, 4, 1 1 2 (or 2A)* 4, 3 5 |
| * Typical race 3 strains are sometimes referred to as biovar 2A. New race 3 strains from the Amazon basin have been placed in a new biovar, designed as 2T or N2 (their relation to races is unclear). | |||
Recently, a new classification scheme has been described for strains of R. solanacearum, based on variation of DNA sequences. Twenty-three sequevars and four phylotypes were identified within the species that broadly reflects the ancestral relationships and geographical origins of the strains [Figure 1. Classification and geographic origins of R. solanacearum strains based on sequence analysis of endoglucanase gene sequences. Numbers indicate sequevars (1 to 23). From Fegan and Prior, 2005.].
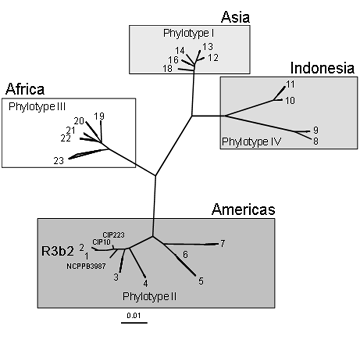
Ralstonia solanacearum race 3 biovar 2 strains belong to phylotype II and sequevars 1 and 2.
Initially described as pathogenic on potato and tomato, R. solanacearum race 3 biovar 2 was shown to also infect and induce symptoms on eggplant, geranium, and pepper. Other solanaceous and non-solanaceous weeds, such as the bittersweet or woody nightshade (Solanum dulcamara) (see the disease cycle and epidemiology section for illustration), are considered as alternate hosts. Most of these alternate hosts remain latently infected and may not show any disease symptoms, but they can be epidemiologically important as inoculum sources and refuges.
Sometimes referred as the ‘cold tolerant’ race, R. solanacearum race 3 biovar 2 originated in the Andes and was probably disseminated worldwide on potato tubers. It is now known to occur in the highlands of the tropics and in subtropical and temperate areas throughout the world, except in North America.
Disease cycle and epidemiology
R. solanacearum race 3 biovar 2 is a soilborne and waterborne pathogen; the bacterium can survive and disperse for various periods of time in infested soil or water, forming a reservoir for source of inoculum for the pathogen. It is not airborne although there was some evidence of survival of the bacterium on the outside of the plant (epiphyte) under conditions of high relative humidity.
The bacterium can survive for days to years in water, wet soils or deep soil layers (>29.5 in / >75 cm), depending on temperature conditions. In aquatic habitats, factors such as pH, salt level, surfaces provided by particulate matter, and the presence of competing, antagonistic or parasitic organisms can affect bacterial survival. Soil moisture content, soil type and plant material content in soil also can play a critical role in its survival in this habitat. At low temperatures (<39.2°F / <4°C), bacterial population densities fall rapidly but the bacteria still can survive, often in a physiological latent state.
In natural habitats, R. solanacerum race 3 biovar 2 can survive during the winter in semi-aquatic weeds, in plant debris or in the rhizophere of non-host plants that act as reservoirs for the pathogen and release bacteria when temperatures start to increase after winter.
R. solanacearum primarily infects host plants through the roots. It penetrates the host through wounds at the points of emergence of lateral roots or through root damage that may be caused by soil-borne microorganisms, such as the root-knot nematode, or by handling. The synergistic interaction between the root-knot nematode and R. solanacearum on a variety of hosts is widely recognized. It can also penetrate into plants by way of stem injuries from insects, handling or tools. Once infection has occurred in the roots, bacteria will colonize the plant through the xylem in the vascular bundles, a process which is accelerated by higher temperatures. R. solanacearum race 3 biovar 2 is most severe on plants between 75-95ºF (24-35°C) (optimal temperature of 80.5ºF-27°C) and decreases in virulence when temperatures exceed 95ºF (35°C) or fall below 53.6ºF (12°C).
Because of vegetative propagation, infected potato seed tubers and infected geranium cuttings can play a major role in dissemination of the bacterium, particularly during latent infections. The pathogen is not known to be disseminated through tomato seeds. Plant-to-plant contamination can occur when bacteria move from roots of infected plants or weeds to roots of nearby healthy plants. The pathogen can be spread from infested to healthy fields by soil transfer on machinery and surface runoff water after irrigation or rainfall. It also can be disseminated from infected ponds or rivers to healthy fields through waterways. In Europe, the semi aquatic weed Solanum dulcamara (bittersweet or climbing nightshade) [Photo 15. Solanum dulcamara (woody nightshade or bittersweet). Key features for identification] infected with R. solanacearum race 3 biovar 2 was shown to play a major role in propagation of the pathogen by releasing bacteria into irrigation water supplies. Recent outbreaks of brown rot of potato in the U. K. have been associated with the use of contaminated river water for irrigation.
In greenhouse production, ebb-and-flow subirrigation systems are conductive to bacterial dissemination and as such are a major risk.
Detection and identification
Detection and identification of R. solanacearum from either symptomatic or asymptomatic plants and from water or soil samples is possible with several microbiological and molecular methods. A battery of complementary tests that differ in their sensitivity and/or specificity should be used for field or laboratory analyses for unambiguous identification of bacteria to genus, species, race and biovar.
Full description of procedures and schemes for R. solanacearum race 3 biovar 2 detection and identification is available in the European Union Council Directive 98-57-EC [http://plantpath.ifas.ufl.edu/rsol/RalstoniaPublications_PDF/Protocols_UE_CoucilDirective_98-57-EC_2006.pdf].
At species level, a number of rapid screening tests can facilitate early detection and identification of R. solanacearum in potentially infected plants or contaminated soil and water samples. However, these tests cannot be used to identify the organism to race or biovar.
The “stem streaming test” might indicate the presence of bacteria in highly infected stems of usually symptomatic potato, geranium and tomato. Bacterial ooze (white spontaneous streaming of bacterial slime) might be observed a few minutes after placing cross-sections of stems into clear water, indicating infestation of vascular bundles by the bacteria [Photo 16. Bacterial streaming in clear water from stem cross-section of plant infected by R. solanacearum].
A common way of confirming the presence of the bacterium in diseased tissue is to isolate the bacterium on a medium that favors bacterial growth. R. solanacearum is relatively easy to isolate from water and soil samples, and plant extracts, using a semi-selective medium, called modified SMSA medium. Typical bacterial colonies appear fluidal, irregular in shape, and white with pink centers after 2 to 5 days incubation at 82.4ºF (28°C) [Photo 17. Appearance of virulent colonies of R. solanacearum on modified SMSA medium].
Several other solid (agar) or liquid (broth) media have been developed for semi-selective detection of R. solanacearum. Plating tests are easy to use and have very good sensitivity.
Immunodiagnostic assays can be used for rapid identification of R. solanacearum from bacterial cultures or symptomatic plant tissue extracts. These tests are based on the ability of specific antibodies to recognize and link to antigens, specific to R. solanacearum. Quick serological tests can be used in the field or greenhouse for early identification of the pathogen. Several quick tests that were evaluated by the USDA-APHIS-PPQ-CPHST Laboratory are available commercially [Photo 18. Result of a quick serological test showing negative (-) and positive (+) detection of R. solanacearum].
Serological methods are generally quick and reliable but suffer from problems with specificity, sensitivity or both. Additionally, they do not distinguish live cells from dead cells.
Several other tests that require minimum equipment can be used for rapid identification of R. solanacearum in the laboratory. Some tests based on differential total fatty acid bacterial composition (Fatty Acid Methyl Ester analysis) and differential utilization of several carbon sources analysis (BIOLOG™ kits) are available commercially and could be used to identify pure cultures of R. solanacearum, but these are expensive and require technical expertise. Serological methods such as ELISA (Enzyme-Linked ImmunoSorbent Assay) and immunofluorescence are relatively inexpensive, easy, fairly fast, and tolerate foreign material in the sample. A number of R. solanacearum-specific nucleic acid-based methods that use the polymerase chain reaction (PCR) amplification can detect both living and dead cells and are more specific and sensitive than serological approaches.
Sensitivity of these methods in complex sample substrates can be significantly improved by using enrichment techniques (in semi-selective growth medium) or DNA purification methods prior to serological detection or PCR amplification.
Pathogenicity tests should be performed as a complementary confirmation for identification of suspect R. solanacearum. These tests consist of assessment of virulence on potato, tomato or tobacco plants.
At the sub-species level, phylotype determination of R. solanacearum can be achieved by multiplex-PCR amplification with different phylotype-specific primer combinations. Several methods can be used for strain characterization, including whole-cell protein profile analysis, genomic fingerprinting or sequence analysis of selected genomic sequences. Most of these techniques are highly reproducible, but require fairly expensive equipment and expertise. The phylotype multiplex PCR followed by sequencing of an internal fragment of the endoglucanase (egl) gene will determine phylotype and sequevar of a strain (see the causal organism section for more details).
A biovar test is used for biovar determination of R. solanacearum. The test is based on the ability of strains of R. solanacearum to differentially produce acid from several carbohydrate sources, including disaccharides and sugar alcohols [Figure 19. Result of a biovar test showing positive (+) and negative (-) utilization of sorbitol by several strains of R. solanacearum].
The test is easy, inexpensive and reproducible. Nevertheless, it should only be used once a strain has already been identified as R. solanacearum using other methods.
Race determination is not generally possible because R. solanacearum strains usually have numerous hosts and do not have race-cultivar specificity on plant hosts. This is why the race sub-classification system has fallen out of favor with scientists, although it still has regulatory meaning because of quarantine rules written for “race 3 biovar 2”.
It is important to understand that unequivocal identification of R. solanacearum race 3 biovar 2 must rely on at least two distinct methods, including the biovar test and one of the nucleid acid-based tests that use PCR to amplify one of several specific DNA fragment.
Currently, for regulatory purposes, the only laboratory with proper registrations for ultimate determination of race and biovar of R. solanacearum race 3 biovar 2 is the USDA-APHIS-PPQ National Plant Germplasm and Biotechnology Laboratory in Beltsville, M. D.
| USDA-APHIS-PPQ-CPHST BARC-East, Bldg. 580 Powder Mill Road Beltsville, MD 20705 Phone number: 301-504-7100 Fax number: 301-504-8539 |
Management
Some level of bacterial wilt control on potato and tomato is possible using resistant cultivars. However, resistance in these host plants may vary across locations, according to variation in temperature. Similarly, the use of antibiotics (streptomycin, ampicillin, tetracycline and penicillin) and soil fumigation has shown little efficacy on R. solanacearum.
As a consequence, there are a number of alternative methods for control of R. solanacearum race 3 biovar 2. In regions where the disease is endemic, cultural control methods appear to be effective in some conditions for reducing bacterial populations of R. solanacearum and subsequent disease incidence: crop rotation can be particularly effective for controlling R. solanacearum race 3 biovar 2, since the pathogen exhibits a fairly narrow host range. The length of time for crop rotation can vary, but rotations of at least two to five years involving several different non-host crops may be used for significant disease reduction; intercropping can be effective for small farmers as cultivation of beans or maize was shown to significantly reduce disease incidence; control of weeds, which have the potential to serve as inoculum reservoirs, in conjunction with crop rotation can also be effective in reducing disease incidence. Control of root-knot nematode populations and cultural practices that minimize root damage can also reduce disease severity.
Some other cultural strategies that might be used to reduce bacterial populations of R. solanacearum race 3 biovar 2 or decrease disease incidence on different hosts include: planting in uninfested production sites, removal of long-term survival sites, selection of appropriate planting and harvest times, deep plowing of crop residues, ensuring satisfactory soil drainage, or early- and late-season irrigation management.
Chemical treatment by soil fumigation has been used to control soil infestation by R. solanacearum race 3 biovar 2, but the method is environmentally destructive, expensive, and difficult to apply, and results are often unsatisfactory. Several different plant essential oils were successfully used as soil fumigants to reduce bacterial populations of R. solanacearum in tomato. Application of stable bleaching powder in conjunction with deep ploughing can also be used as it showed significant reduction in bacterial populations of the pathogen in greenhouse and field trials in several geographic areas. However, soil disinfection appears to be soil dependent and not universally applicable. Modification of soil pH by use of acidified nutrient solution or a combination of organic amendment and fertilizers was shown to be very effective in reducing bacterial wilt diseases on diverse hosts. Heat treatment by solarization when used in combination with other control strategies is another method that was shown to reduce R. solanacearum populations in soil. Efficacy of heat treatment on control of the bacterium may vary according to soil moisture content, heat temperature and duration of heat application. Application of plant resistance inducers, such as acibenzolar-S-methyl, might be used to enhance host resistance against R. solanacearum race 3 biovar 2, as it was recently shown to work for broad-host range strains of the pathogen. On geranium, application of phosphorous acid as a drench was recently shown to protect host plants from infection by the bacterium. Finally, use of suppressive soils was shown to slow infection of tomato seedlings by R. solanacearum and reduce bacterial wilt incidence in nurseries.
Initial studies on biological control of R. solanacearum gave promising results. The use of numerous diverse bacterial strains antagonistic to R. solanacearum as control agents gave positive results in the greenhouse or in strictly controlled field tests. Similarly, antagonists that are closely related to or derived from the wild type of R. solanacearum itself, such as spontaneous or genetically engineered avirulent mutants of the bacterium, were shown to confer protection against bacterial wilt disease on potato and tomato under greenhouse conditions. Such antagonists may be able to colonize and survive asymptomatically on the host without reducing yield. The use of these strains as antagonist agents for control of R. solanacearum under true field conditions has to be evaluated.
In the United States and other areas where R. solanacearum race 3 biovar 2 is not known to be established, the first strategy is to prevent introduction and inadvertent spread of the pathogen. This can be achieved by the establishment of exclusionary and sanitary practices, along with governmental regulations.
A “New Pest Response Guidelines” (USDA-APHIS-PPQ) [http://www.aphis.usda.gov/import_export/plants/manuals/emergency/downloads/nprg-ralstonia.pdf] and a "Recovery plan for Ralstonia solanacearum race 3 biovar 2" (USDA-ARS) [http://www.ars.usda.gov/SP2UserFiles/Place/00000000/opmp/Rs3-2RecoveryPlan-v-Oct112006.pdf] give the most accurate available information for detection, control, containment, and eradication of R. solanacearum race 3 biovar 2.
In areas where the pathogen is present but not yet established everywhere, it is critically important to observe good cultural sanitation practices to keep uninfested areas clean. These include planting only certified disease-free seed potatoes or cuttings, disinfesting all equipment before moving from field to field, and never using surface water for irrigation. Even where R. solanacearum race 3 biovar 2 is present in soils (as in many parts of the African highlands), use of disease-free seed potatoes can significantly reduce disease incidence and allow growers to harvest a profitable crop. Effective health management of brown rot of potatoes, Southern wilt of geranium and bacterial wilt of tomato caused by R. solanacearum race 3 biovar 2 will be possible through establishment of specific strategies and practices adapted for each of these diseases.
A “Mimimum sanitation protocols for offshore geraniums cutting production” document [http://www.aphis.usda.gov/plant_health/plant_pest_info/ralstonia/downloads/ralstoniaworkplan.pdf] was developed by APHIS-PPQ in 2004 to be used by off-shore geranium suppliers. It defines minimum standards and requirements for greenhouse structure and material.
Regulation
In the United States, R. solanacearum race 3 biovar 2 is listed as a Select Agent, a designation under the Plant Protection Act (7 CFR Part 330) and the Agricultural Bioterrorism Protection Act of 2002 (7 CFR Part 331) that defines the possession, use, and transfer of Select Agents and Toxins.
State diagnostic laboratories receiving suspect infected plant material or cultures are required to have permits from the USDA-APHIS-Plant Protection and Quarantine (consult the PPQ permit website at http://www.aphis.usda.gov/plant_health/permits/index.shtml). Laboratories possessing, using, or transferring Select Agents such as R. solanacearum race 3 biovar 2 are required to be registered with the PPQ. Diagnostic laboratories that are not registered and identify R. solanacearum race 3 biovar 2, or receive positive feedback from the USDA Laboratory, from a suspect sample are required to immediately notify the APHIS Select Agent Program within seven calendar days, and either destroy or transfer the agent to a registered laboratory within seven days.
References
Allen, C., Kelman, A., and French, E. R. 2001. Brown rot. Pages 11-13 in: Compendium of potato diseases, 2nd. ed. Stevenson, W. R., Loria, R., Franc, G. D., and Weingartner, D. P., eds. APS Press, St. Paul, M. N.
Autrique, A. and Potts, M. 1987. The influence of mixed cropping on the control of potato bacterial wilt (Pseudomonas solanacearum). Annals of Applied Biology 111:125-133.
Boshou, L. 2005. A broad review and perspective on breeding for resistance to bacterial wilt. Pages 225-238 in: Bacterial wilt disease and the Ralstonia solanacearum species complex. Allen, C., Prior, P., and Hayward, A. C., eds. APS press, St. Paul, M. N.
CABI/EPPO. 1999. Distribution maps of plant diseases. Map N0. 785 CAB International. Wallingford, U. K.
Caruso, P., Gorris, M. T., Cambra, M., Palomo, J. L., Collar, J., and Lopez, M. M. 2002. Enrichment double-antibody sandwich indirect enzyme-linked immunosorbent assay that uses a specific monoclonal antibody for sensitive detection of Ralstonia solanacearum in asymptomatic potato tubers. Applied and Environmental Microbiology 68:3634-3638.
Cook, D., Barlow, E., and Sequeira, L. 1989. Genetic diversity of Pseudomonas solanacearum: Detection of restriction fragment length polymorphisms with DNA probes that specify virulence and the hypersensitive response. Molecular Plant-Microbe Interactions 2:113-121.
Cook, D. and Sequeira, L. 1991. The use of subtractive hybridization to obtain a DNA probe specific for Pseudomonas solanacearum race 3. Molecular and General Genetics 227:401-410.
Cook, D. and Sequeira, L. 1994. Strain differentiation of Pseudomonas solanacearum by molecular genetic methods. Pages 77-93 in: Bacterial wilt: the disease and its causative agent, Pseudomonas solanacearum. A. C. Hayward and G. L. Hartman, eds. CAB International, Wallingford, U. K.
Coutinho, T. A. 2005. Introduction and prospectus on the survival of R. solanacearum. Pages 29-38 in: Bacterial wilt disease and the Ralstonia solanacearum species complex. Allen, C., Prior, P., and Hayward, A. C., eds. APS press, St. Paul, M. N.
Denny, T. P. 2006. Plant pathogenic Ralstonia species. Pages 573-644 in: Plant-associated bacteria. S. S. Gnanamanickam, ed. Springer Publishing, Dordrecht, The Netherlands.
Denny, T. P. and Hayward, A. C. 2001. Gram-negative bacteria: Ralstonia. Pages 151-174 in: Laboratory guide for identification of plant pathogenic bacteria, 3rd ed. Schaad, N. W., Jones, J. B., and Chun, W., eds. APS Press, St. Paul, M. N.
Douglas, S. M. 2002. Diseases of geranium. The Connecticut Agricultural Experiment Station, New Haven, C. T.
Elphinstone, J. G. 2001. Monitoring and control of the potato brown rot bacterium (Ralstonia solanacearum) in the UK: A case study in: Proceedings of the FNK/EAPR/ESA/UEITP 2nd European Potato Processing Conference. Held Nov., 14-15th. Lausanne, Switzerland.
Elphinstone, J. G. 2005. The current bacterial wilt situation: a global overview. Pages 9-28 in: Bacterial wilt disease and the Ralstonia solanacearum species complex. C. Allen, P. Prior, and A. C. Hayward, eds. APS press, St-Paul, M. N.
Elphinstone, J. G., Henessy, J., Wilson, J. K., and Stead, D. 1996. Sensitivity of different methods for the detection of Ralstonia solanacearum in potato tuber extracts. Bulletin OEPP/EPPO Bulletin 26:663-678.
Elphinstone, J. and Harris, L. 2002. Monitoring and control of the potato brown rot bacterium in irrigation water. 2 p. British Potato Council, Oxford, U. K.
Englebrecht, M. C. 1994. Modification of a semi-selective medium for the isolation and quantification of Pseudomonas solanacearum. Pages 3-5 in: Bacterial wilt newsletter. Hayward, A. C., ed. ACIAR, Canberra, Australia.
Farag, N. S., Lashin, S. M., All-Abdel, R. S., Shatta, H. M., and Seif-Elyazal, A. M. 1982. Antibiotics and control of potato black leg and brown rot diseases. Agricultural Research Review 60:149-166.
Fegan M. and Prior P. 2005. How complex is the "Ralstonia solanacearum" complex ? Pages 449-461 in: Bacterial wilt disease and the Ralstonia solanacearum species complex. C., Allen, P., Prior, A. C., Hayward, eds. APS Press, St. Paul, M. N.
Fegan, M., Holoway, A. C., Hayward, A. C., and Timmis, J. 1998. Development of a diagnostic test based on the polymerase chain reaction to identify strains of R. solanacearum exhibiting the biovar 2 genotype. Pages 34-43 in: Bacterial wilt disease: Molecular and ecological aspects. P., Prior, C., Allen, and J., Elphinstone, eds. Springer Verlag, Berlin, Germany.
Floyd, J. 2003. Action plan for Ralstonia solanacearum race 3, biovar 2 found in nursery facilities. 23 p. Published Feb., 27, Version 3. USDA, APHIS, PPQ. Pest Detection and Management Programs, Riverdale, M. D.
French, E. R., Gutarra, L., Aley, P., and Elphinstone, J. 1995. Culture media for Pseudomonas solanacearum isolation, identification and maintenance. Fitopatologia 30:126-130.
Graham, J. and Lloyd, A. B. 1978. Solanum cinereum R. Br., a wild host of Pseudomonas solanacearum biotype II. Journal of the Australian Institute of Agricultural Science 44:124-126.
Grey, B. E. and Steck, T. R. 2001. The viable but nonculturable state of Ralstonia solanacearum may be involved in long-term survival and plant infection. Applied and Environmental Microbiology 67:3866-3872.
Hayward, A. C. 1975. Biotypes of Pseudomonas solanacearum in Australia. Australian Plant Pathology Society Newsletter 4:9-11.
Hayward, A. C. 1991. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annual Review of Phytopathology 29:65-87.
Janse, J. D. 1996. Potato brown rot in western Europe - history, present occurrence and some remarks on possible origin, epidemiology and control strategies. Bulletin OEPP/EPPO Bulletin 26:679-695.
Janse, J. D., van den Beld, H. E., Elphinstone, J., Simpkins, S., Tjou-Tam-Sin, L. N. A., and van Vaerenbergh, J. 2005. Introduction to Europe of Ralstonia solanacearum biovar 2, race 3 in Pelargonium zonale cuttings from Kenya. Pages 81-94 in: Bacterial wilt disease and the Ralstonia solanacearum species complex. C. Allen, P. Prior, and A. C. Hayward, eds. APS Press, St-Paul, M. N.
Kelman, A. 1954. The relationship of pathogenicity of Pseudomonas solanacearum to colony appearance in a tetrazolium medium. Phytopathology 44:693-695.
Kelman, A. and Sequeira, L. 1965. Root-to-root spread of Pseudomonas solanacearum. Phytopathology 55:304-309.
Lambert, C. D. 2002. Agricultural bioterrorism protection act of 2002: possession, use, and transfer of biological; agents and toxins; interim and final rule. (7 CFR Part 331). Federal Register 67:76908-76938.
Lemaga, B., Kanzikwera, R., Kakuhenzire, R., Hakiza, J. J., and Manzi, G. 2001. The effect of crop rotation on bacterial wilt incidence and potato tuber yields. African Crop Sciencey Journal 9:257-266.
Lemay, A., Redlin, S., Fowler, G., and Dirani, M. 2003. Pest data sheet: Ralstonia solanacearum race 3 biovar 2. Published Feb., 12. USDA-APHIS-PPQ. Center for plant health science and technology. Plant epidemiology and risk analysis laboratory, Raleigh, N. C.
Luo, K. and Wang, Z. 1983. Study of bacterial wilt (Pseudomonas solanacearum) controlled by antagonistic and avirulent P. solanacearum. Acta Phytopathological Sinica 13:51-56.
McCarter, S. M. 1991. Bacterial wilt. Pages 28-29 in: Compendium of tomato diseases. Jones, J. B., Jones, J. P., Stall, R. E., and Zitter, T. A., eds. APS Press, St. Paul, M. N.
Mclaughlin, R. J. and Sequeira, L. 1988. Evaluation of an avirulent strain of Pseudomonas solanacearum for biological control of bacterial wilt of potato. American Potato Journal 65:255-268.
Michel, V. V. and Mew, T. W. 1998. Effect of a soil amendment on the survival of Ralstonia solanacearum in different soils. Phytopathology 88:300-305.
Nishiyama, M., Shiomi, Y., Suzuki, S., and Marumoto, T. 1999. Suppression of growth of Ralstonia solanacearum, tomato bacterial wilt agent, on/in tomato seedlings cultivated in a suppressive soil. Soil Science and Plant Nutrition 45:79-87.
Norman, D. J., Chen, J., Yuen, J. M. F., Mangravita-Novo, A., Byrne, D., and Walsh, L. 2006. Control of bacterial wilt of geranium with phosphorous acid. Plant Disease 90:798-802.
Olsson, K. 1976. Experience of brown rot caused by Pseudomonas solanacearum. EPPO Bulletin 6:199-207.
Persley, G. J. 1986. Ecology of Pseudomonas solanacearum, the causal agent of bacterial wilt. Pages 71-76 in: Bacterial wilt disease in Asia and the South Pacific. ACIAR Proceedings N0. 13. G. J. Persley, ed. ACIAR, Canberra, Australia.
Pradeep, K. and Sood, A. K. 2001. Integration of antagonistic rhizobacteria and soil solarization for the management of bacterial wilt of tomato caused by Ralstonia solanacearum. Indian Phytopathology 54:12-15.
Pradhanang, P. M., Ji, P., Momol, M. T., Olson, S. M., and Jones, J. B. 2005. Application of acibenzolar-S-methyl enhances host resistance in tomato against Ralstonia solanacearum. Plant Disease 89:989-993.
Pradhanang, P.M., Momol, M.T., Olson, S.M., and Jones, J.B. 2003. Effects of plant essential oils on Ralstonia solanacearum population density and bacterial wilt incidence in tomato. Plant Disease 87:423-427.
Priou, S., Gutarra, L., and Aley, P. 2006. An improved enrichment broth for the sensitive detection of Ralstonia solanacearum (biovars 1 and 2A) in soil using DAS-ELISA. Plant Pathology 55:36-45.
Rowe, R. C. and Powelson, M. L. 2007. Potato health management: a holistic approach. Pages 1-5 in: Potato health management, Second ed. Johnson, D. A., ed. APS Press Publisher: St. Paul, M.N.
Roy, S., Ojha, P. K., Ojha, K. L., Upadhyay, J. P., and Jha, M. M. 1999. Effect of mixed application of fertilizers and organic amendments on the disease intensity of wilt complexes on banana (Musa sp.). Journal of Applied Biology 9:84-86.
Saddler, G. S. 2005. Management of bacterial wilt disease. Pages 121-132 in: Bacterial wilt disease and the Ralstonia solanacearum species complex. Allen, C., Prior, P., and Hayward, A. C., eds. APS press, St. Paul, M. N.
Schaad, N. W., Gaush, P. E., Ozakman M., inventors. 2007. Real-time PCR primers and probes for identification of Ralstonia solanacearum race 3, biovar 2 in potato and other plants. Issued: Aug., 28. Patent 6,410,223.
Smith, J. J. and Saddler, G. S. 2001. The use of avirulent mutants of Ralstonia solanacearum to control bacterial wilt disease. Pages 159-176 in: Biotic interactions in plant-pathogen associations. Jeger, M. J. and Spence, N. J., eds. CABI Publishing, Wallingford, U. K.
Smith, J. J., Offord, L. C., Holderness, M., and Saddler, G. S. 1995. Genetic diversity of Burkholderia solanacearum (synonym Pseudomonas solanacearum) race 3 in Kenya. Applied and Environmental Microbiology 61:4262-4268.
Stefani, E. and Mazzucchi, U. 1997. Protein electrophoretograms for the identification of Ralstonia solanacearum in potato tubers. Journal of plant pathology 79:189-195.
Swanson, J. K., Yao, J., Tans-Kersten, J., and Allen, C. 2005. Behavior of Ralstonia solanacearum race 3 biovar 2 duing latent infection of geranium. Phytopathology 95:136-143.
Trigalet, A., Trigalet-Demery, D., and Prior, P. 1998. Elements of biocontrol of tomato bacterial wilt. Pages 332-336 in: Bacterial wilt disease: Molecular and ecological aspects. Prior, P., Allen C., and Elphinstone J., eds. Springer-Verlag, Berlin, Germany.
Van der Wolf, J. M., Bonants, P. J. M., Smith, J. J., Hagenaar, M., Nijhuis, E., van Beckhoven, J. R. C., Saddler, G. S., Trigalet, A., and Feuillade, R. 1998. Genetic diversity of Ralstonia solanacearum race 3 in Western Europe as determined by AFLP, RC-PFGE and Rep-PCR. Pages 44-49 in: Bacterial wilt disease: Molecular and ecological aspects. P., Prior, C., Allen, and J., Elphinstone, eds. Springer Verlag, Berlin, Germany.
Van Elsas, J.D., Kastelein, P., van Bekkum, P., van der Wolf, J.M., de Vries, P.M., and van Overbeek, L.S. 2000. Survival of Ralstonia solanacearum biovar 2, the causative agent of potato brown rot, in field and microcosm soils in temperate climates. Phytopathology 90:1358-1366.
Weller, S. A., Elphinstone, J. G., Smith, N. C., Boonham, N., and Stead, D. E. 2000. Detection of Ralstonia solanacearum strains with a quantitative, multiplex, real-time, fluorogenic PCR (TaqMan) assay. Applied and Environmental Microbiology 66:2853-2858.
Yabuuchi, E., Kosako, Y., Yano, I., Hotta, H., and Nishiuchi, Y. 1995. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov.: Proposal of Ralstonia pickettii (Ralston, Palleroni and Douderoff 1973) comb. nov., Ralstonia solanacearum (Smith 1896) comb. nov. and Ralstonia eutropha (Davis 1969) comb. nov. Microbiology and Immunology 39, 897-904.
Yao, G., Zhang, F., and Li, Z. 1994. Control of bacterial wilt with soil amendment. Chinese Journal of Biological Control 10:106-109.
Glossary
(In order of appearance in the text)
Pathogen. A pathogen, or infectious agent, is a biological agent that causes disease or illness to its host. A number of different organisms can cause plant infectious disease. Among them are fungi, bacteria, viruses, nematodes or parasitic plants.
Bacterium. A bacterium is a microscopic organism consisting of individual cells. Bacteria cause diseases in many host plants. They can survive on crop residue, seed, or in soil and water; they may be spread by plant or plant cuttings transfer, mechanical means, insects, and seeds.
Symptoms. A symptom is a subjective evidence of disease or physical disturbance. It is an evident reaction by a plant to a pathogen, and is not necessarily visible. Different pathogens can induce similar symptoms.
Vascular bundles. Vascular, or conductive, bundles are responsible for long-distance transport of water and nutrients throughout the plant. Highly developed plants have two types of vascular tissues: the xylem and the phloem.
Stolons. A stolon is a slender stem that grows horizontally along the ground, giving rise to roots and aerial (vertical) branches at specialized points called nodes.
Chlorosis. Chlorosis is a condition in which leaves produce insufficient chlorophyll. As chlorophyll is responsible for the green color of leaves, chlorotic leaves are pale, yellow, or yellow-white.
Necrotic. In plant biology, necrosis is the name given to death of plant cells and plant tissue. The tissue first turns brown and subsequently dies.
Xylem. The xylem is responsible for transportation of raw sap (water and nutrients) from roots to aerial parts of the plant. R. solanacearum is a limited xylem-invading pathogen.
Solanaceae. The Solanaceae family, also known as the "nightshade" family, is a family of flowering plants, many of which are edible, while others are poisonous. The family includes the Datura or Jimson weed, eggplant, mandrake, deadly nightshade or belladonna, capsicum, potato, tobacco, tomato, and petunia.
Gram-negative. Bacteria have been classified as Gram-negative or Gram-positive regarding structural differences in their cell walls. Many species of Gram-negative bacteria are pathogenic. This pathogenic capability is usually associated with certain components of Gram-negative cell walls.
Aerobic. An aerobic organism requires oxygen for aerobic cellular respiration. Cellular respiration is the mechanism by which aerobic organism require oxygen to utilize substrates (for example sugars and fats) in order to obtain energy.
Bacterial colonies. On the surface of a solid growth medium, individual bacterial cells will grow and multiply to become visible bacterial colonies. All cells within the colony descend from a single ancestor and are identical. Characteristics of bacterial colonies (color, aspect, diameter or growth rate) are commonly used for bacteria identification.
Virulent. Virulence refers to the degree of pathogenicity of a microorganism, or in other words the relative ability of a microorganism to cause disease.
Selective medium. A growth or culture medium is a substance in which microorganisms, such as bacteria, or cells can grow. Selective media are used for the growth of only select microorganisms. They usually contain antibiotics to which the select microorganims is resistant to.
Strains. A strain is a genetic variant or subtype of a microorganism (for example virus or bacterium or fungus).
Variability. Variability here refers to variation of a given characteristic from one bacterial strain (or group of strains) to the other.
Species. A species is one of the basic units of biological classification. A species is often defined as a group of organisms capable of interbreeding and producing fertile offspring. Each species is placed within a single genus.
Species complex. R. solanacearum is considered a "species complex" as it includes individual isolates that may not be considered within a single species, as it is the case for the banana blood disease bacterium or Pseudomonas syzygii.
Carbohydrate substrates. Carbohydrates are simple organic compounds such as sugars and starch which contain carbon chains. They fill numerous roles in living organisms, such as the storage and transport of energy and structural components. Carbohydrates are differentially used as source of energy by bacteria.
Races. A race is formed by a group of bacterial strains that are differentiated based on the response on a set of host differentials.
Biovars. A biovar is a group of bacterial strains that are distinguishable from other strains of the same species on the basis of their physiological characteristics.
Variation of DNA sequences. Comparison of DNA sequences is commonly used for classification studies of strains of microorganisms. It is basically assumed that the higher the homology is between two strains, the more closely related the strains are in terms of evolution. These types of studies are known as phylogenetic studies.
Sequevars. A sequevar, or sequence variant, is defined as a group of strains with a highly conserved sequence within the area sequenced.
Phylotypes. A phylotype is defined as a group of strains that are closely related based on phylogenetic analysis of sequence data. Each phylotype is composed of a number of sequevars.
Endoglucanase. Endoglucanase is a type of cellulase which is a class of enzymes produced by fungi, bacteria, and protozoans that catalyze the cellulolysis (or hydrolysis) of cellulose.
Root-knot nematode. Root-knot nematodes are plant-parasitic roundworms from the genus Meloidogyne. They exist in soil in areas with hot climates or short winters. There are a great many parasitic forms, including pathogens in most plants, animals, and also in humans.
Vegetative propagation. Vegetative propagation, reproduction or multiplication is a type of asexual reproduction found in plants. It is a process by which new plant "individuals" arise or are obtained without production of seeds.
Ebb-and-flow subirrigation. Ebb-and-flow subirrigation is a method of irrigation that is usually used for potted plants in greenhouses. Water is delivered to the plant root zone from below the soil surface and absorbed upwards.
Immunodiagnostic assays. These assays are based on the use of antibiotics in various test formats to detect and identify any molecules or cells (including bacteria). The most commonly used assays for bacteria detection and identification are agglutination, enzyme-linked immunosorbent assay (ELIZA), immunofluorescence, lateral flow strip tests and flow-through assays.
Antibodies. Antibodies (also known as immunoglobulins) are proteins that are used by the immune system to identify and neutralize foreign objects, such as bacteria and viruses. Due to their specificity, they are commonly used in biology for detection and identification of microorganisms.
Antigens. An antigen is any molecule that is recognized by an antibody.
Nucleic-acid. A nucleic acid is a molecule composed of nucleotide chains. These molecules carry genetic information. The most common nucleic acids are deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). Nucleic acids are universal in living things, as they are found in all cells and viruses.
Polymerase chain reaction (PCR) amplification. The polymerase chain reaction is a technique that consists of amplifying a DNA molecule exponentially.
Multiplex-PCR. In multiplex-PCR amplification, multiple primer pairs are used for DNA amplification.
Cultivars. A cultivar is a cultivated plant that has been selected and given a unique name because it has desirable characteristics (decorative or useful) that distinguish it from otherwise similar plants of the same species.
Endemic. Endemic, in a broad sense, can mean "belonging" or "native to", "characteristic of", or "prevalent in" a particular geography, area, or environment; native to an area or scope.
Disease incidence. Disease Incidence is a measure of the risk of developing the disease within a specified period of time
Plant resistance inducers. Plant resistance inducers are natural or synthetic chemical compounds that apparently act by stimulating the natural defense response in the plant.
Suppressive soils. A suppressive soil is one that possesses some level of control of a disease forming organism. All soils have a natural level of disease suppressive activities. In most soils long term management can either reduce or increase this level of suppression.
Biological control. Biological control is defined as the reduction of pest populations (including insects, mites, weeds and plant diseases) by natural enemies. Biological control agents of plant diseases are most often referred to as antagonists.
This R. solanacearum/Bacterial wilt - R3b2 page has been optimized for print. To view this page in its original form, please visit: http://plantpath.ifas.ufl.edu/rsol/Trainingmodules/RalstoniaR3b2_PrinterText.html
©2008 R. solanacearum race 3 biovar 2 USDA-NRI project. All rights reserved.