| Ralstonia solanacearum Race 3 Biovar 2 Descalgar la guía descriptiva al formato PDF |
|||
| Roll your mouse over the red-colored words to see word definitions. This will open Pop-up windows. If not, print the glossary |
|||
| Printer friendly |
PDF file |
Glossary |
|
| Symptoms & signs | Causal organism | Disease cycle & epidemiology | Detection & identification | Management | References |
| Detection and identification |
Detection and identification of R. solanacearum from either symptomatic or asymptomatic plants and from water or soil samples is possible with several microbiological and molecular methods. A battery of complementary tests that differ in their sensitivity and/or specificity should be used for field or laboratory analyses for unambiguous identification of bacteria to genus, species, race and biovar.
Full description of procedures and schemes for R. solanacearum race 3 biovar 2 detection and identification is available in the European Union Council Directive 98-57-EC.
At species level, a number of rapid screening tests can facilitate early detection and identification of R. solanacearum in potentially infected plants or contaminated soil and water samples. However, these tests cannot be used to identify the organism to race or biovar.
The “stem streaming test” might indicate the presence of bacteria in highly infected stems of usually symptomatic potato, geranium and tomato. Bacterial ooze (white spontaneous streaming of bacterial slime) might be observed a few minutes after placing cross sections of stems into clear water, indicating infestation of vascular bundles by the bacteria (Photo 16). |
 |
Photo 16. Bacterial streaming in clear water from stem cross-section |
A common way of confirming the presence of the bacterium in diseased tissue is to isolate the bacterium on a medium that favor bacterial growth. R. solanacearum is relatively easy to isolate from water and soil samples, and plant extracts using a semi-A growth or culture medium is a substance in which microorganisms, such as bacteria, or cells can grow. Selective media are used for the growth of only select microorganisms. They usually contain antibiotics to which the select microorganims is resistant toselective medium, called modified SMSA medium. Typical bacterial colonies appear fluidal, irregular in shape, and white with pink centers after 2 to 5 days incubation at 28ºC82.4ºF (Photo 17). |
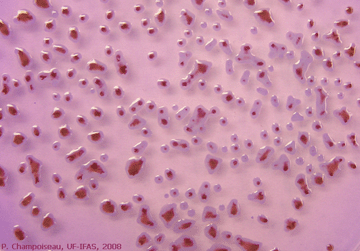 |
Photo 17. Appearance of virulent colonies of R. solanacearum |
These assays are based on the use of antibiotics in various test formats to detect and identify any molecules or cells (including bacteria). The most commonly used assays for bacteria detection and identification are agglutination, enzyme-linked immunosorbent assay (ELIZA), immunofluorescence, lateral flow strip tests or flow-through assaysImmunodiagnostic assays can be used for rapid identification of R. solanacearum from bacterial cultures or symptomatic plant tissue extracts. These tests are based on the ability of specific
Antibodies (also known as immunoglobulins) are proteins that are used by the immune system to identify and neutralize foreign objects, such as bacteria and viruses. Due to their specificity, they are commonly used in biology for detection and identification of microorganismsantibodies to recognize and link to
An antigen is any molecule that is recognized by an antibodyantigens, specific to R. solanacearum. Quick serological tests can be used in the field or greenhouse for early identification of the pathogen. Several quick tests that were evaluated by the USDA-APHIS-PPQ-CPHST Laboratory are available commercially (Photo 18).
Serological methods are generally quick and reliable but suffer from problems with specificity, sensitivity or both. Additionally, they do not distinguish live cells from dead cells.
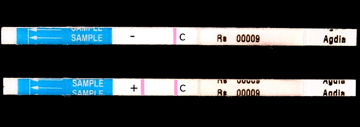 |
Photo 18. Result of a quick serological test showing negative (-) |
Several other tests that require minimum equipment can be used for rapid identification of R. solanacearum in the laboratory. Some tests based on differential total fatty acid bacterial composition (Fatty Acid Methyl EsterFAME analysis) and differential utilization of several carbon sources analysis (BIOLOG™ kits) are available commercially and could be used to identify pure cultures of R. solanacearum, but these are expensive and require technical expertise. Serological methods such as
Enzyme-Linked ImmunoSorbent AssayELISA and immunofluorescence are relatively inexpensive, easy, fairly fast, and tolerate foreign material in the sample. A number of R. solanacearum-specific
A nucleic acid is a molecule composed of nucleotide chains. These molecules carry genetic information. The most common nucleic acids are deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). Nucleic acids are universal in living things, as they are found in all cells and virusesnucleic-acid-based methods that use the
The polymerase chain reaction is a technique that consists of amplifying a DNA molecule exponentiallypolymerase chain reaction (PCR) amplification can detect both living and dead cells and are more specific and sensitive than serological approaches.
Sensitivity of these methods in complex sample substrates can be significantly improved by using enrichment techniques (in semi-selective growth medium) or DNA purification methods prior to serological detection or PCR amplification.
Pathogenicity tests should be performed as a complementary confirmation for identification of suspect R. solanacearum. These tests consist of assessment of virulence on potato, tomato or tobacco plants.
At the sub-species level,
A phylotype is defined as a group of strains that are closely related based on phylogenetic analysis of sequence data. Each phylotype is composed of a number of sequevarsphylotype determination of R. solanacearum can be achieved by
In multiplex-PCR amplification, multiple primer pairs are used for DNA amplificationmultiplex-PCR amplification with different phylotype-specific primer combinations. Several methods can be used for strain characterization, including whole-cell protein profile analysis, genomic fingerprinting or sequence analysis of selected genomic sequences. Most of these techniques are highly reproducible, but require fairly expensive equipment and expertise. The phylotype multiplex PCR followed by sequencing of an internal fragment of the endoglucanase (egl) gene will determine phylotype and sequevar of a strain (see the causal organism section for more details).
A biovar test is used for biovar determination of R. solanacearum. The test is based on the ability of strains of R. solanacearum to differentially produce acid from several carbohydrate sources, including disaccharides and sugar alcohols (Figure 19). |
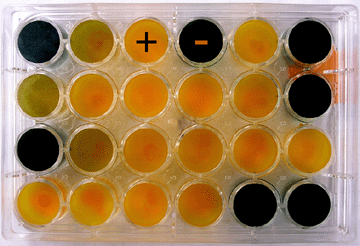 |
Photo 19. Result of a biovar test showing positive (+) and negative (-) |
Race determination is not generally possible because R. solanacearum strains usually have numerous hosts and do not have race-cultivar specificity on plant hosts. This is why the race sub-classification system has fallen out of favor with scientists, although it still has regulatory meaning because of quarantine rules written for “race 3 biovar 2”.
It is important to understand that unequivocal identification of R. solanacearum race 3 biovar 2 must rely on at least two distinct methods, including the biovar test and one of the nucleid acid-based tests that use PCR to amplify one of several specific DNA fragment.
Currently, for regulatory purposes, the only laboratory with proper registrations for ultimate determination of race and biovar of R. solanacearum race 3 biovar 2 is the USDA-APHIS-PPQ National Plant Germplasm and Biotechnology Laboratory in Beltsville, M. D.
| USDA-APHIS-PPQ-CPHST BARC-East, Bldg. 580 Powder Mill Road Beltsville, MD 20705 Phone number: 301-504-7100 Fax number: 301-504-8539 |
| Symptoms & signs | Causal organism | Disease cycle & epidemiology | Detection & identification | Management | References |
| Author: | Patrice G. Champoiseau of University of Florida |
| Reviewers: | Caitilyn Allen of University of Wisconsin; Jeffrey B. Jones, Carrie Harmon and Timur M. Momol of University of Florida |
| Publication date: | September 12, 2008 |
| Supported by: | The United States Department of Agriculture - National Research Initiative Program (2007-2010) |

